1:
2:
3:
4:
5:
6:
7:
8:
9:
10:
11:
12:
13:
14:
15:
16:
17:
18:
19:
20:
21:
22:
23:
24:
25:
26:
27:
28:
29:
30:
31:
32:
33:
34:
35:
36:
37:
38:
39:
40:
41:
42:
43:
44:
45:
46:
47:
48:
49:
50:
51:
52:
53:
54:
55:
56:
57:
58:
59:
60:
61:
62:
63:
64:
65:
66:
67:
68:
69:
70:
71:
72:
73:
74:
75:
76:
77:
78:
79:
80:
81:
82:
83:
84:
|
############################################################
# Data
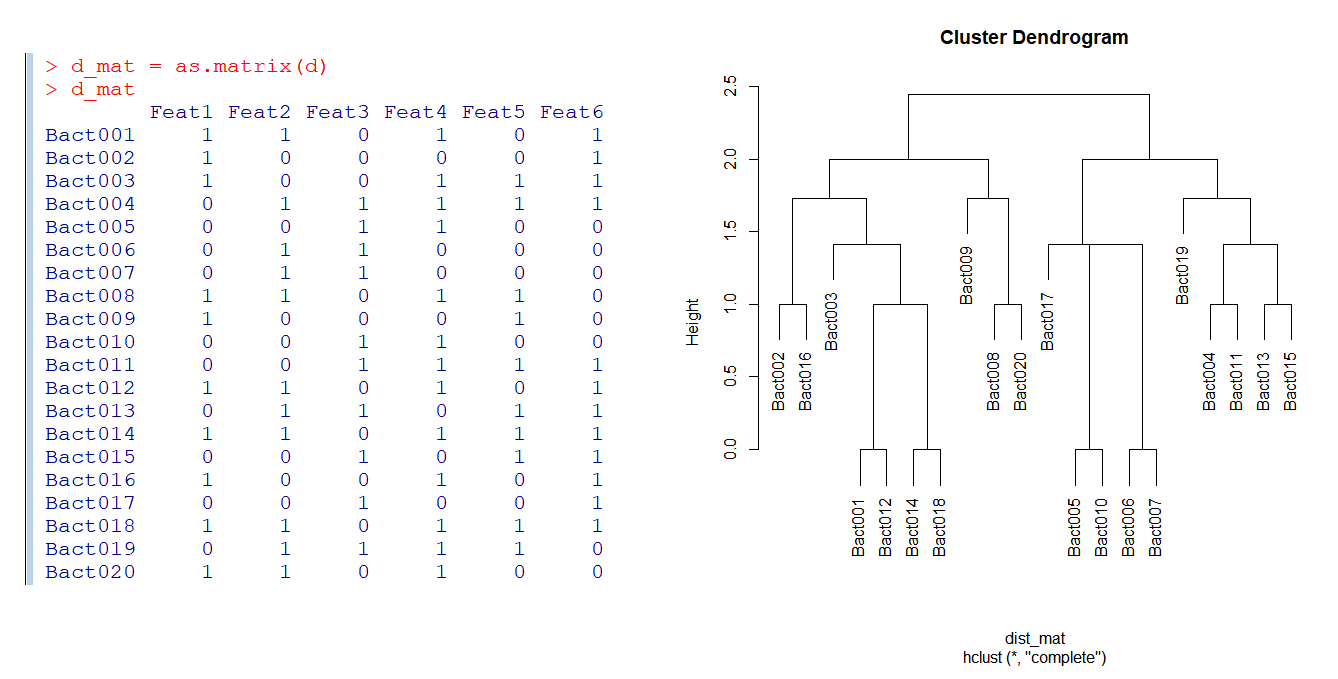
d = read.table(header = TRUE, text =
"Feat1 Feat2 Feat3 Feat4 Feat5 Feat6
Bact001 1 1 0 1 0 1
Bact002 1 0 0 0 0 1
Bact003 1 0 0 1 1 1
Bact004 0 1 1 1 1 1
Bact005 0 0 1 1 0 0
Bact006 0 1 1 0 0 0
Bact007 0 1 1 0 0 0
Bact008 1 1 0 1 1 0
Bact009 1 0 0 0 1 0
Bact010 0 0 1 1 0 0
Bact011 0 0 1 1 1 1
Bact012 1 1 0 1 0 1
Bact013 0 1 1 0 1 1
Bact014 1 1 0 1 1 1
Bact015 0 0 1 0 1 1
Bact016 1 0 0 1 0 1
Bact017 0 0 1 0 0 1
Bact018 1 1 0 1 1 1
Bact019 0 1 1 1 1 0
Bact020 1 1 0 1 0 0
")
d_mat = as.matrix(d)
d_mat
############################################################
# Simple Dendrogram
dist_mat = dist(d_mat, method="euclidean") # distance matrix (diagonal)
dist_mat
dist_clust = hclust(dist_mat) # clustering based on distance
dist_clust
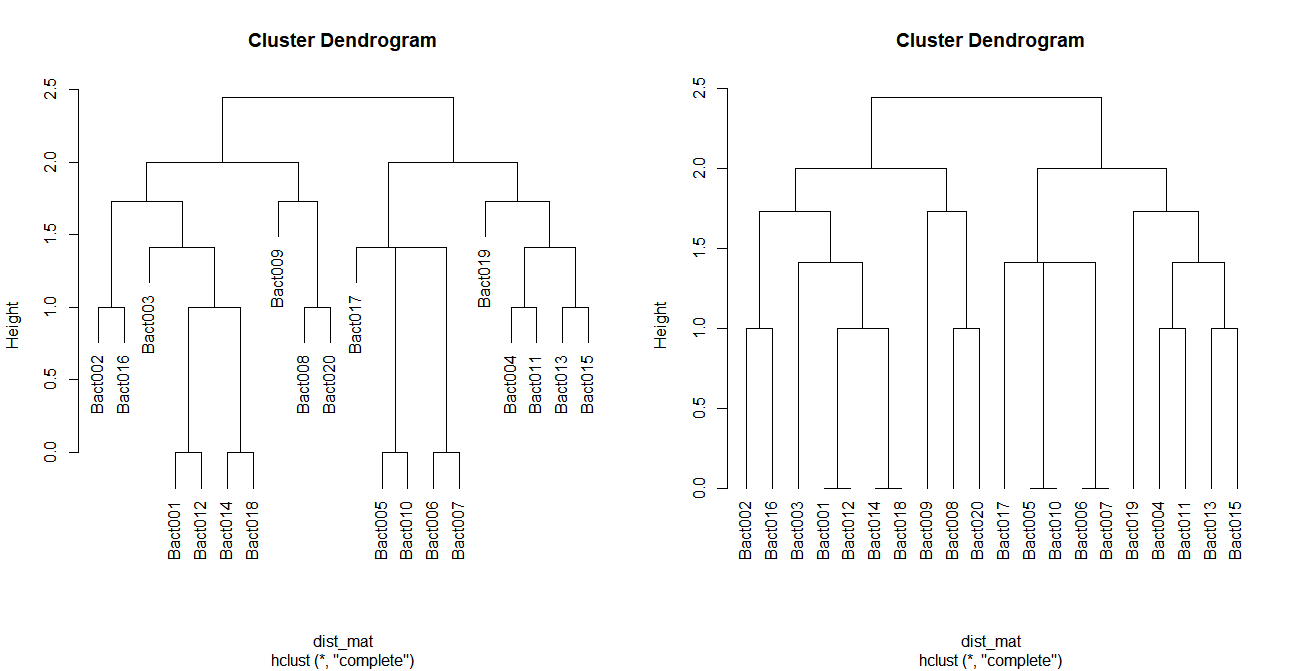
par(mfrow=c(1, 2))
plot(dist_clust, cex = 1)
plot(dist_clust, cex = 1, hang = -1)
############################################################
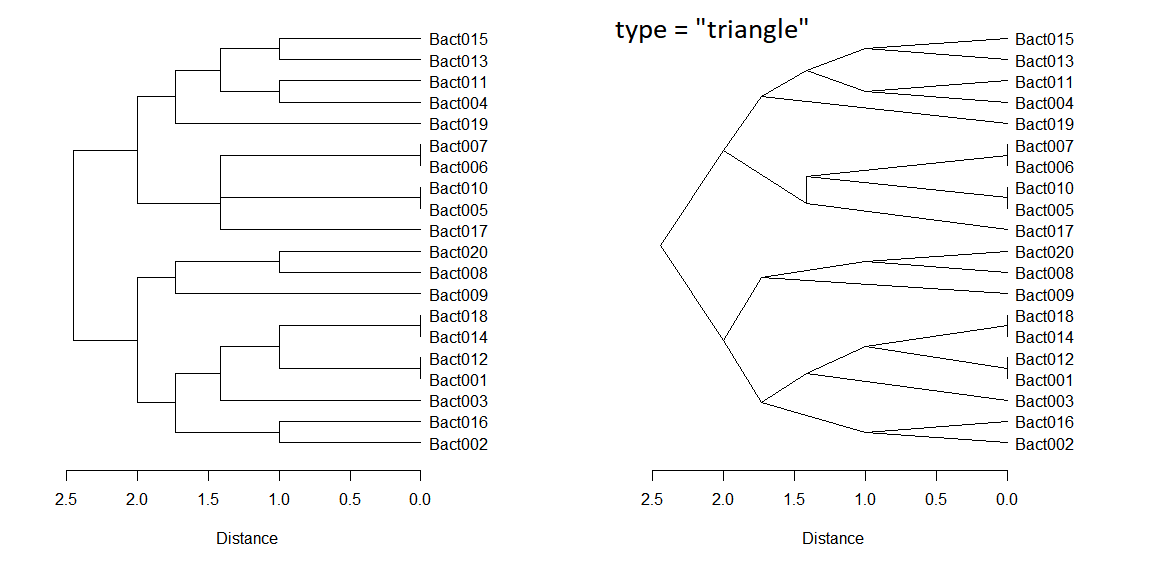
par(mfrow=c(1, 2)); par(mar=c(8, 3, 3, 8))
dend = as.dendrogram(dist_clust)
plot(dend, cex = 1, horiz = TRUE, xlab="Distance")
plot(dend, cex = 1, horiz = TRUE, xlab="Distance", type = "triangle")
############################################################
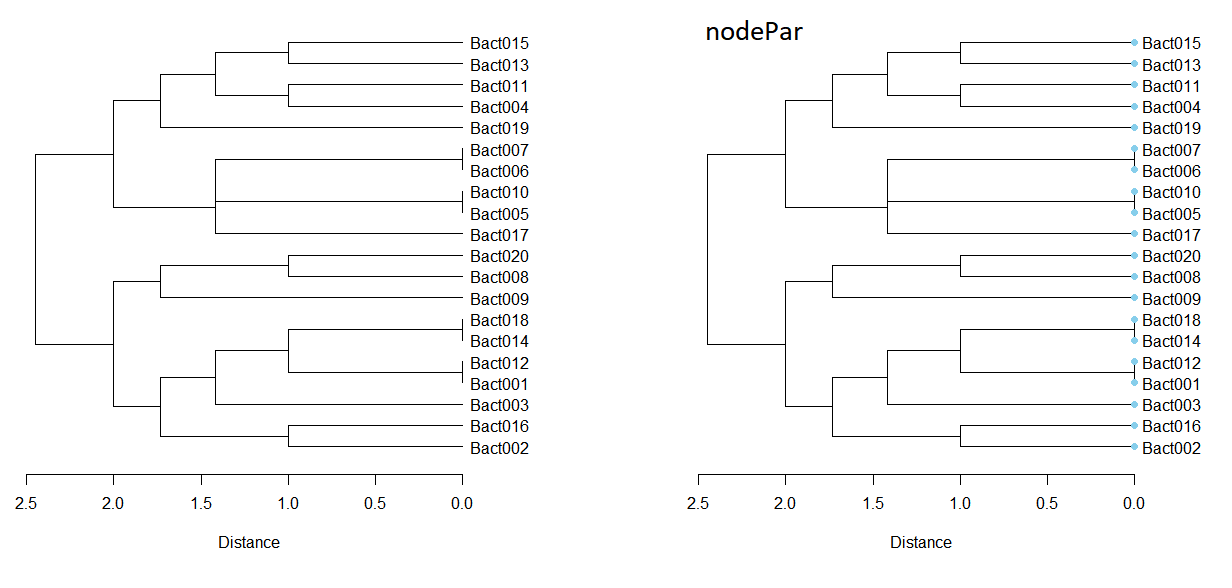
par(mfrow=c(1, 2)); par(mar=c(8, 3, 3, 8))
plot(dend, cex = 1, horiz = TRUE, xlab="Distance")
nodeSet = list(lab.cex = 1.0, pch = c(NA, 19), cex = 1.0, col = "skyblue")
plot(dend, cex = 1, horiz = TRUE, xlab="Distance", nodePar = nodeSet)
############################################################
# From dendrogram to phylogenetic tree
# install.packages("ape")
library("ape")
tree = as.phylo(dist_clust)
tree$edge.length
par(mfrow=c(1, 2)); par(mar=c(1,1,1,1))
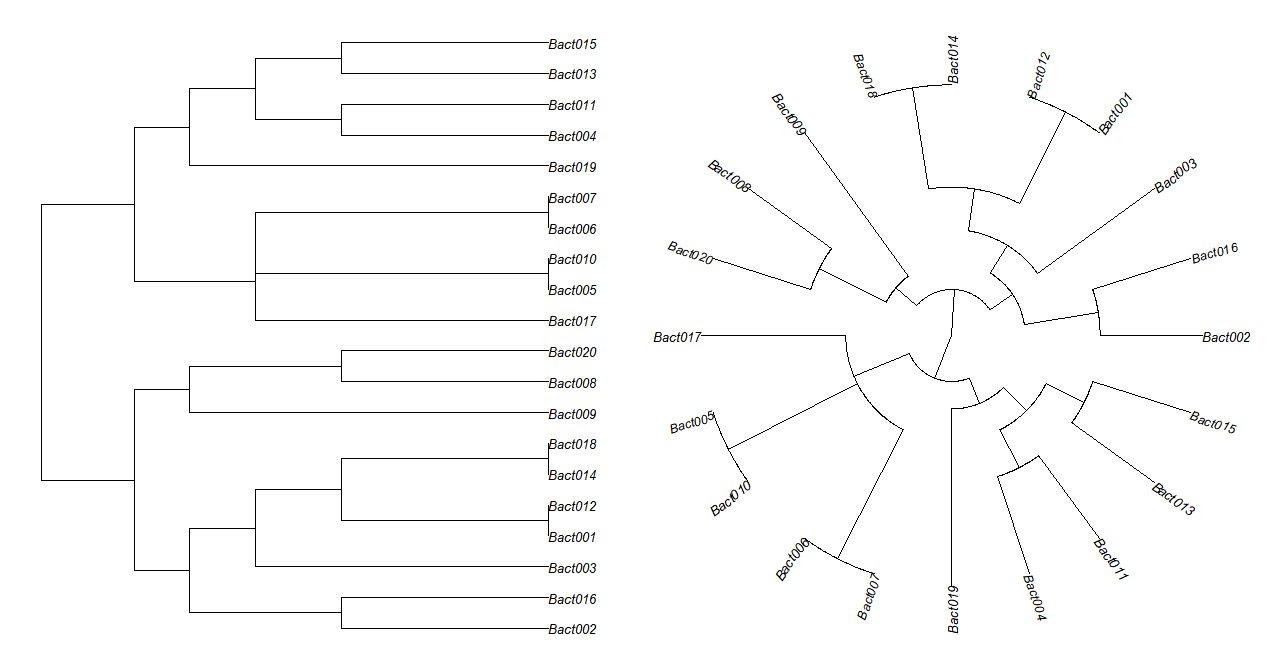
# "phylogram", "cladogram", "fan", "unrooted", "radial"
plot(tree, cex=0.8, type = "phylogram")
plot(tree, cex=0.8, type = "fan")
############################################################
lab=tree$tip.label
lab
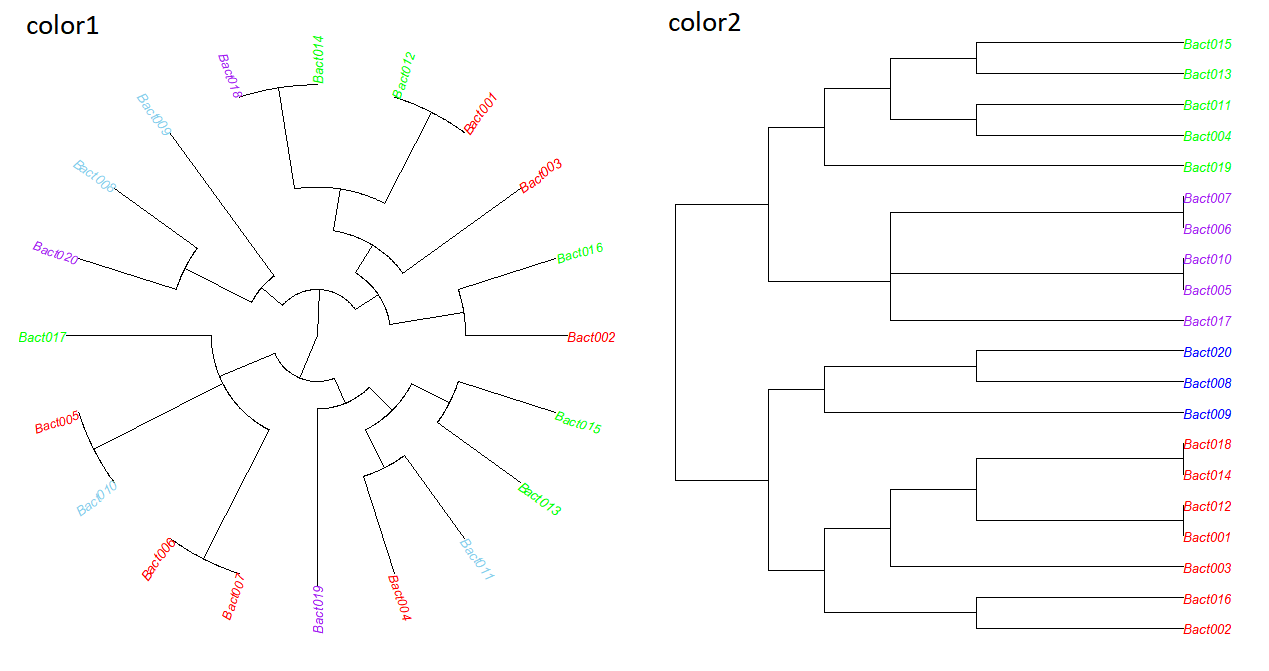
color1 = c("red", "red", "red", "red", "red", "red", "red",
"skyblue", "skyblue", "skyblue", "skyblue",
"green", "green", "green", "green", "green", "green",
"purple", "purple", "purple")
color2 = c("red", "red", "red", "green", "purple", "purple", "purple",
"blue", "blue", "purple", "green", "red", "green", "red",
"green", "red", "purple", "red", "green", "blue")
cbind(lab, color1)
cbind(lab, color2)
plot(tree, cex=0.8, tip.color=color1, type = "fan")
plot(tree, cex=0.8, tip.color=color2)
# For more information
http://www.sthda.com/english/wiki/beautiful-dendrogram-visualizations-in-r-5-must-known-methods-unsupervised-machine-learning
http://www.r-graph-gallery.com/31-custom-colors-in-dendrogram/
|