1:
2:
3:
4:
5:
6:
7:
8:
9:
10:
11:
12:
13:
14:
15:
16:
17:
18:
19:
20:
21:
22:
23:
24:
25:
26:
27:
28:
29:
30:
31:
32:
33:
34:
35:
36:
37:
38:
39:
40:
41:
42:
43:
44:
45:
46:
47:
48:
49:
50:
51:
52:
53:
54:
55:
56:
57:
58:
59:
60:
61:
62:
63:
64:
65:
66:
67:
68:
69:
70:
71:
72:
73:
74:
75:
76:
77:
78:
79:
80:
81:
82:
83:
84:
85:
86:
87:
88:
89:
90:
91:
92:
93:
94:
95:
96:
97:
98:
99:
100:
|
# ANOVA using a Vector Object
x = c(5.9, 6.7, 5.3, 6.4, 0.4, 0.3, 0.3, 0.3, 1.8, 1.4, 1.2, 0.8)
x
maxX=max(x)*1.2
groups = c(rep("A Samples", 4), rep("B Samples", 4), rep("C Samples", 4))
g=factor(groups)
colors = c("blue", "red", "green")
d=data.frame(x, g)
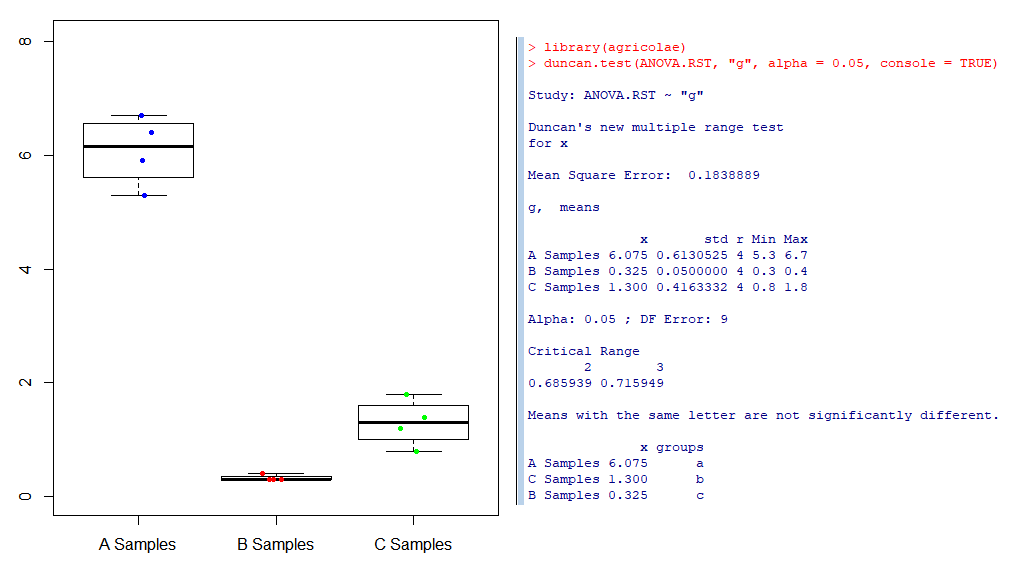
boxplot(x~g, data=d, ylim=c(0,maxX))
# Statistical properties of data shown in box plot are as follows
# mean (open square), box (25/75% quantile),
# whisker (5/95% quantile), and asterisks (maximum/minimum).
stripchart(x~g, data=d,
vertical = TRUE, method = "jitter",
pch = 20, col = colors, bg = "bisque",
add = TRUE)
# ANOVA and Multiple Comparison - Tukey
ANOVA.RST <- aov(x ~ g)
x_aov = anova(ANOVA.RST)
summary(ANOVA.RST)
summary(x_aov)
TukeyHSD(ANOVA.RST)
# ANOVA and Multiple Comparison - Duncan
install.packages("agricolae")
library(agricolae)
duncan.test(ANOVA.RST, "g", alpha = 0.05, console = TRUE)
# ANOVA and Multiple Comparison - Graph with Letters
# install.packages("multcomp")
library(multcomp)
x = c(5.9, 6.7, 5.3, 6.4, 0.4, 0.3, 0.3, 0.3, 1.8, 1.4, 1.2, 0.8)
x
maxX=max(x)*1.2
groups = c(rep("Group_A", 4), rep("Group_B", 4), rep("Group_C", 4))
g=factor(groups)
colors = c("blue", "red", "green")
d=data.frame(x, g)
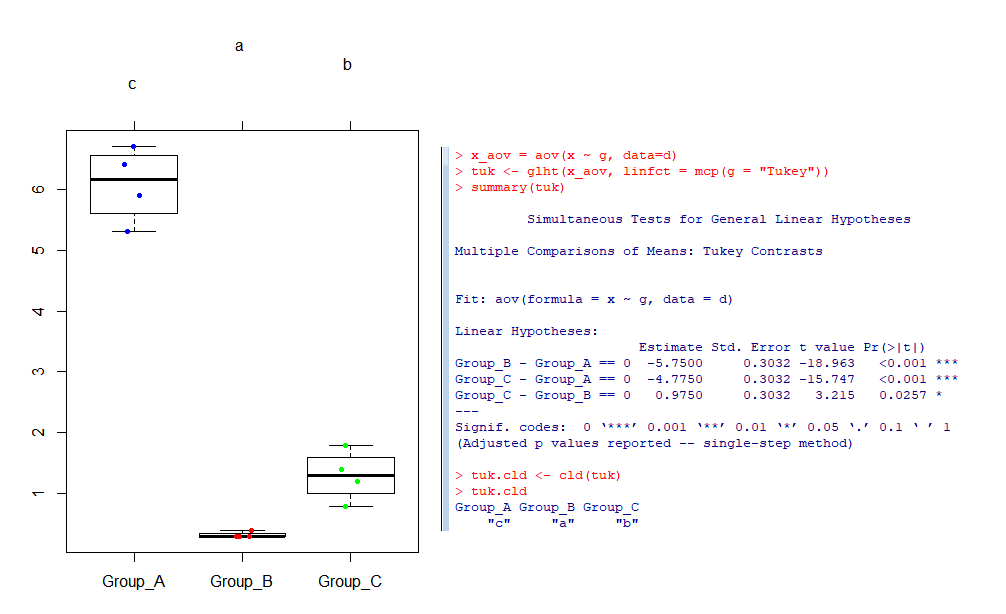
x_aov = aov(x ~ g, data=d)
s=summary(x_aov)
s
lm = aov(x ~ g, data=d)
anova(x_aov)
anova(lm)
x_aov_mean = model.tables(x_aov,"means")
x_aov_mean
as.numeric(x_aov_mean[[1]]$g)
anova(x_aov)
anova_p = s[[1]][1, 5];
anova_p
multiComp <- glht(x_aov, linfct = mcp(g = "Tukey"))
confint(multiComp)
plot(print(confint(multiComp)))
summary(multiComp) # For Tukey's Test
summary(multiComp, test=univariate()) # For repeated-t and Fisher's LSD Test
summary(multiComp, test=adjusted(type="bonferroni")) # Bonferroni's Test
p.adjust.methods
# Refernce: http://www.math.montana.edu/jobo/st541/mcp.r
# Refernce: https://www.rdocumentation.org/packages/agricolae/versions/1.2-8/topics/LSD.test
multiComp.cld <- cld(multiComp)
multiComp.cld
multiComp.cld <- cld(multiComp, level=0.1)
multiComp.cld
summary(multiComp.cld)
multiComp.cld[[10]]
multiComp.cld[[10]]$Letters
as.character(multiComp.cld[[10]]$Letters)
mean_by_group = tapply(x, g, mean)
mean_by_group
as.numeric(mean_by_group)
sd_by_group = tapply(x, g, sd)
sd_by_group
as.numeric(sd_by_group)
par(oma = c(2, 2, 2, 2)) # spaces for all sides.
par(mar = c(2, 2, 7, 2)) # spaces for all sides.
plot(multiComp.cld)
#box("inner" , col="gray" , lty=12) # "plot", "figure", "inner" and "outer"
stripchart(x~g, data=d,
vertical = TRUE, method = "jitter",
pch = 20, col = colors, bg = "bisque",
add = TRUE)
|