1:
2:
3:
4:
5:
6:
7:
8:
9:
10:
11:
12:
13:
14:
15:
16:
17:
18:
19:
20:
21:
22:
23:
24:
25:
26:
27:
28:
29:
30:
31:
32:
33:
34:
35:
36:
37:
38:
39:
40:
41:
42:
43:
44:
45:
46:
47:
48:
49:
|
library(rgl)
library(devtools)
install_github("vqv/ggbiplot")
library(ggbiplot)
# PCA
pcadata <- princomp(iris[,1:4], cor=T, scores=T)
# cor = T: Use the correlation matrix or the covariance matrix
# score = T: each principal component calculated!
# summary
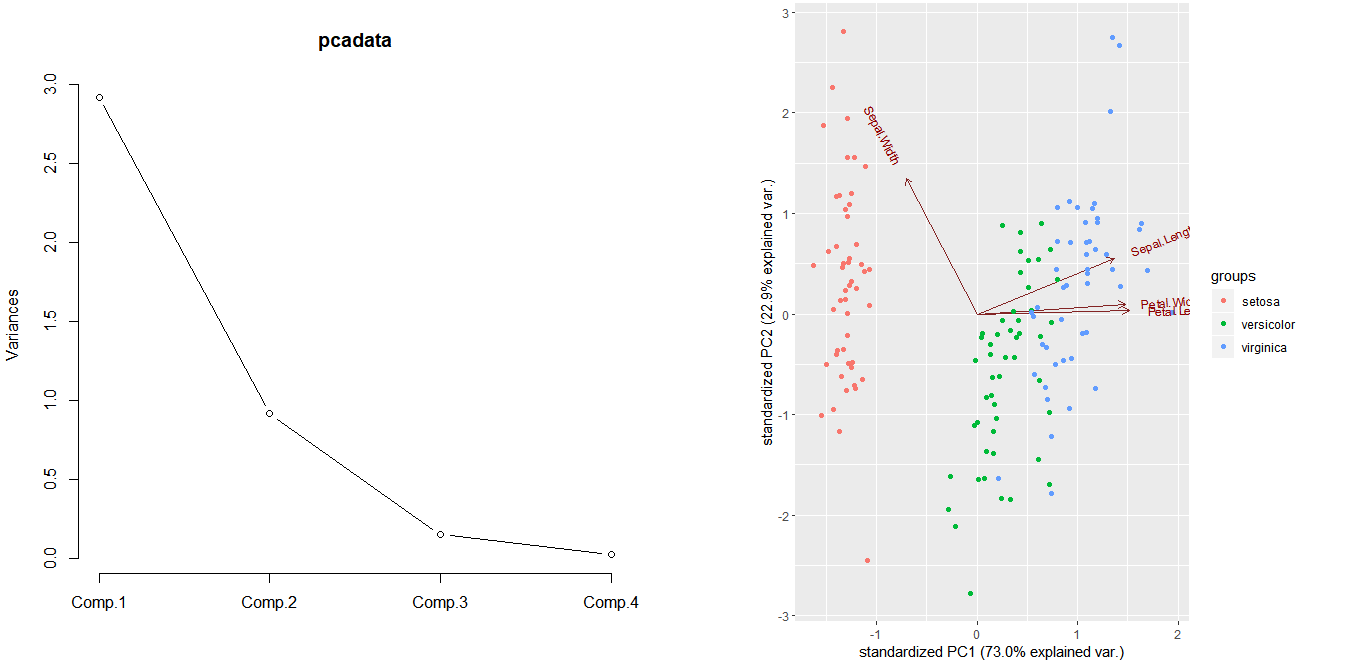
plot(pcadata, type='l')
summary(pcadata)
# 2d score plot
ggbiplot(pcadata,groups = iris$Species)
# loading plot
plot(pcadata$loadings[,1:2], col=c('black','blue','red','green'), pch=16)
legend('topleft',
c('Sepal.Length','Sepal.Width','Petal.Length','Petal.Width'),
text.col=c('black','blue','red','green'))
install.packages("devtools") # also need install ggplot2
library("devtools")
install_github("ggbiplot")
library("ggbiplot")
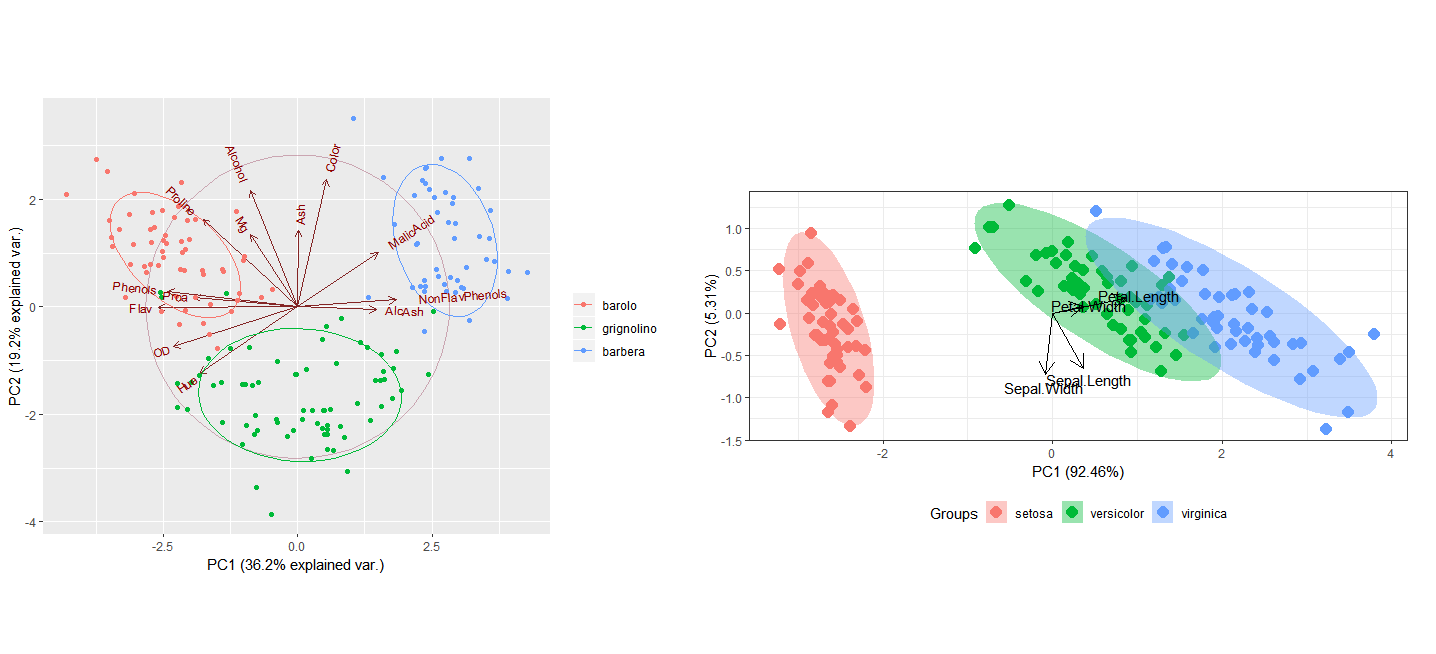
data(wine)
wine.pca <- prcomp(wine,scale.=TRUE)
g<-ggbiplot(wine.pca, obs.scale=1, var.scale=1, groups=wine.class, ellipse=TRUE, circle=TRUE)
g<-g+scale_color_discrete(name="")
print(g)
library(devtools)
install_github('fawda123/ggord')
library(ggord)
ord <- prcomp(iris[, 1:4])
p <- ggord(ord, iris$Species)
p
p + scale_colour_manual('Species', values = c('purple', 'orange', 'blue'))
p + theme_classic()
p + theme(legend.position = 'bottom')
p + scale_x_continuous(limits = c(-4, 4))
|