1:
2:
3:
4:
5:
6:
7:
8:
9:
10:
11:
12:
13:
14:
15:
16:
17:
18:
19:
20:
21:
22:
23:
24:
25:
26:
27:
28:
29:
30:
31:
32:
33:
34:
35:
36:
37:
38:
39:
40:
41:
42:
43:
44:
45:
|
############################################################
# Bar Plot with Error Bars
par(mfrow=c(1,1))
sd.p = function(x) {
m = mean(x);
ss = 0;
for(i in 1:length(x)) {
ss = ss + (x[i] - m)^2;
}
return(sqrt(ss/length(x)));
}
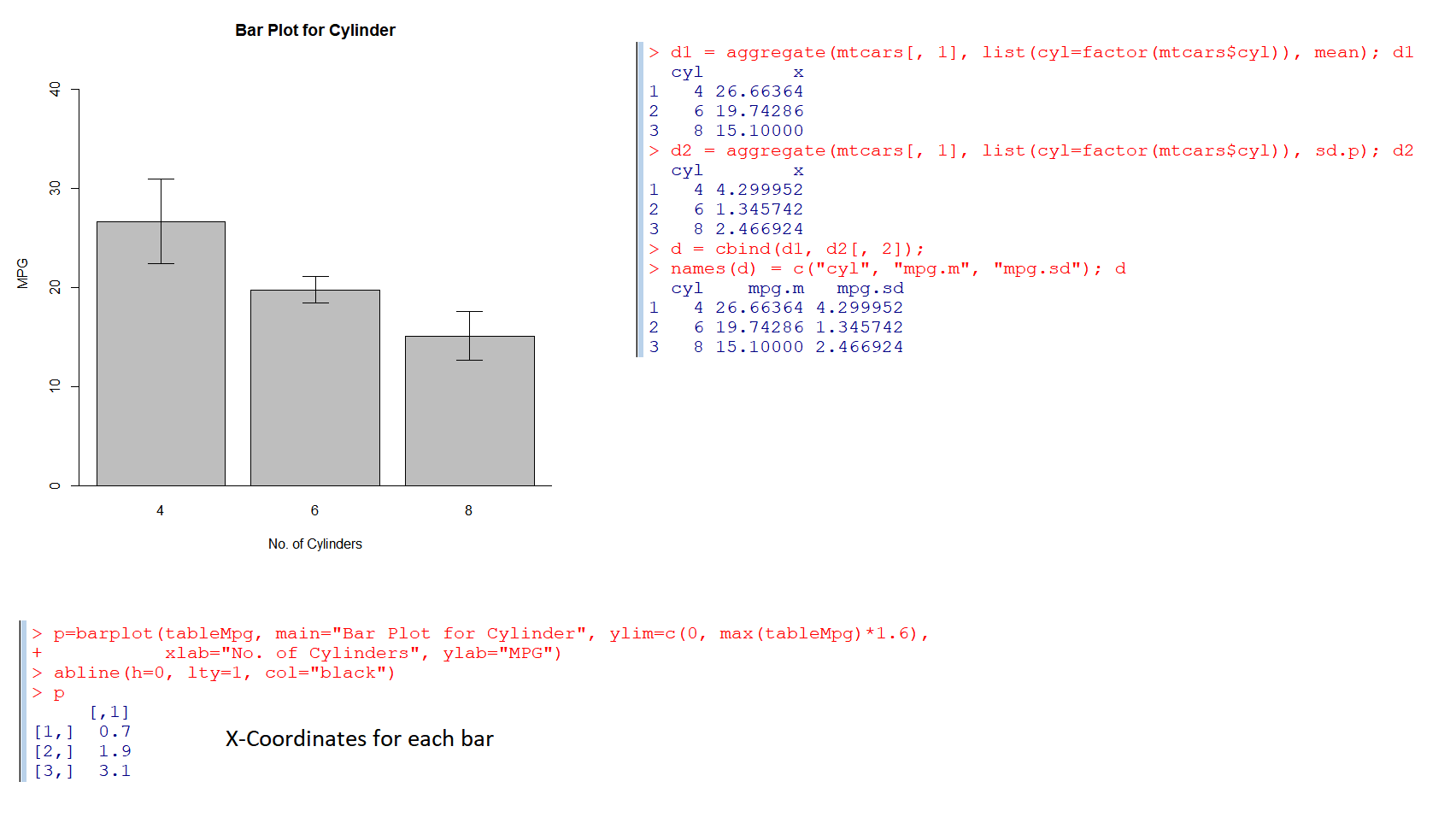
d1 = aggregate(mtcars[, 1], list(cyl=factor(mtcars$cyl)), mean); d1
d2 = aggregate(mtcars[, 1], list(cyl=factor(mtcars$cyl)), sd.p); d2
d = cbind(d1, d2[, 2]);
names(d) = c("cyl", "mpg.m", "mpg.sd"); d
tableMpg = structure(d$mpg.m, .Dim = 3, .Dimnames = structure(list(
d1$cyl), .Names = "Cylinders"), class = "table")
tableMpg
p=barplot(tableMpg, main="Bar Plot for Cylinder", ylim=c(0, max(tableMpg)*1.6),
xlab="No. of Cylinders", ylab="MPG")
abline(h=0, lty=1, col="black")
p
funcErrorBars = function(x, y, xAdd, yAdd) {
x.y_verti = list(x=c(x, x), y=c(y-yAdd, y+yAdd));
lines(x.y_verti, lty=1, col="black");
x.y_horiz1 = list(x=c(x-xAdd, x+xAdd), y=c(y+yAdd, y+yAdd));
lines(x.y_horiz1, lty=1, col="black");
x.y_horiz2 = list(x=c(x-xAdd, x+xAdd), y=c(y-yAdd, y-yAdd));
lines(x.y_horiz2, lty=1, col="black");
}
sd.y = d$mpg.sd
y = d$mpg.m
d
for(i in 1:3) {
funcErrorBars(p[,1][i], y[i], 0.1, sd.y[i]);
}
|